Mar. 16, Thu, 2023
Topics
>RESEARCH
Finding ‘Vault’: Unravelling the Mysteries of p62-Bodies and the Cellular Recycling Pathway
Scientists use a novel technique to identify a protein complex degraded by autophagy with possible role in liver cancer
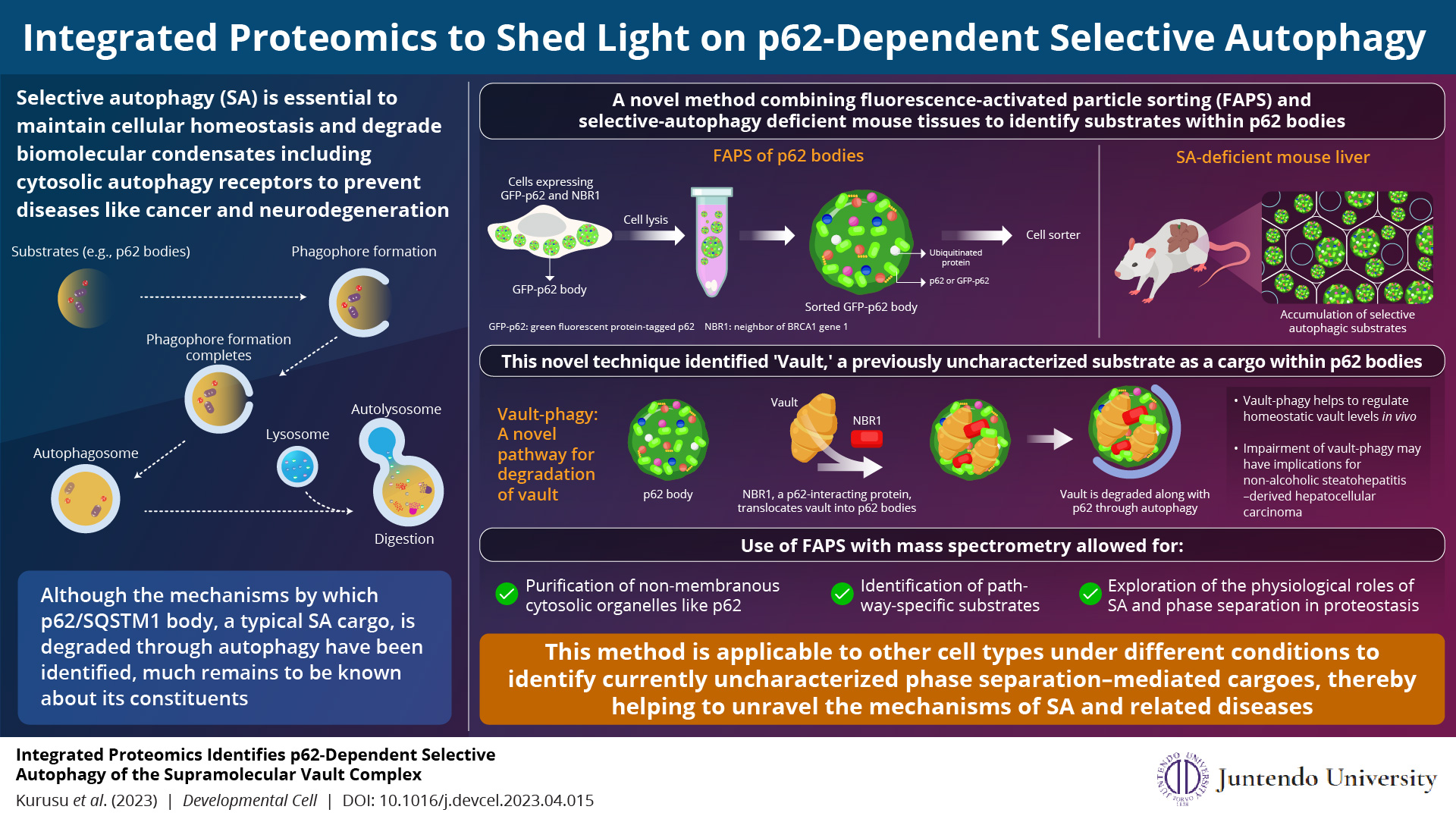 The method led to the identification of vault, a component of p62 bodies, with a possible role innon-alcoholic steatohepatitis-derived hepatocellular carcinoma.
The method led to the identification of vault, a component of p62 bodies, with a possible role innon-alcoholic steatohepatitis-derived hepatocellular carcinoma.
Image credits:Masaaki Komatsu from Juntendo University School of Medicine
License type:Original content
Our body functioning is delicately balanced between the synthesis and breakdown of various cellular components. When these cellular components grow old or get damaged, they are digested by a process called “autophagy”—literally, “self-eating.” This process
not only helps in the elimination of toxic wastes, but also helps to deliver building blocks for the synthesis of new cellular macromolecules. Thus, autophagy serves as the body’s cellular cleaning and recycling system.
Researchers have long been studying the mechanisms related to autophagy and its role in potentially preventing and fighting diseases. p62 protein is an important part of this process. It forms a membrane-less cytosolic organelle called p62 body and helps in the selective removal of toxic cellular waste through autophagy. Regulated removal of unwanted proteins via selective autophagy of p62 bodies maintains cellular homeostasis. However, their compromised clearance leads to the accumulation of p62 and proteins inside p62 bodies, resulting in several diseases, such as hepatocellular carcinoma. While studies have helped us understand the mechanisms associated with the degradation of p62 bodies, our knowledge on their constituents remains incomplete.
To this end, a group of scientists led by Associate Professor Hideaki Morishita and Professor Masaaki Komatsu from Juntendo University Graduate School of Medicine, Japan, have developed a novel method that uses fluorescence-activated particle sorting (FAPS) to purify p62 bodies. They followed it with mass spectrometry of the purified organelles and the tissues retrieved from selective autophagy-defective mice, to identify the substrates within p62 bodies. Their analyses not only identified several previously known autophagy substrates and receptors present within p62 bodies, but also unraveled a major subunit of the supramolecular protein complex, vault, as a cargo within the membrane-less organelle. Their findings have been published in the journal Developmental Cell on May 15, 2023.
The team also included Reo Kurusu and Yuki Fujimoto, undergraduate students from the Program for Training Physicians Conducting Basic Research, Juntendo University. The scientists elaborate that vault is recruited to p62 bodies and degraded by selective autophagy, a process called vault-phagy. This is achieved through an intermediary protein called NBR1, which is critical for the employment of vault and its subsequent degradation along with p62 bodies.
Why is this finding important? According to the scientists, vault-phagy, i.e., the degradation of vault, regulates homeostatic vault levels within cells and tissues, and its impairment may be associated with non-alcoholic steatohepatitis-derived hepatocellular carcinoma, among others.
Discussing their study, Prof. Masaaki remarks, “Our proteomic approach identified the contents of p62 bodies in detail and enabled us to determine the substrates of p62-mediated selective autophagy. This method can be applied to other cell types and tissues under several conditions, like stress and diseases.”
Although there is evidence in the literature on methods to identify selective autophagy substrates, the present method using FAPS and selective autophagy–defective mice has several advantages. FAPS is more suitable for the purification of non-membranous cytosolic organelles like p62 bodies. Furthermore, it can identify pathway-specific substrates and can be instrumental in understanding the physiological role of selective autophagy and phase separation in proteostasis.
As Associate Professor Hideaki Morishita observes, “Our methods have a broad scope of potential applications that should lead to the discovery of currently uncharacterized phase separation–mediated cargoes in order to allow for a deeper understanding of selective autophagy and related diseases.”
While the scientists believe there is a need for further research to understand the implications of vault-phagy in diseases, their novel technique and the insights generated in this study are sure to enrich our knowledge of the mechanisms of our cell’s housekeeping!
Funding
This study is mainly supported by Grant-in-Aid for Scientific Research on Innovative Areas (19H05706), a Grant-in-Aid for Scientific Research (A) (21H004771), Advanced Research and Development Programs for Medical Innovation (AMED-CREST, 22gm1410004s0103), the Takeda Science Foundation, a Grant-in-Aid for Scientific Research (B) (20H03213, 23H02441), Advanced Research and Development Programs for Medical Innovation (AMED-PRIME, 21gm6410019h0001), the Inamori Scientific Foundation (217959), a Kishimoto Fund Research Grant (2022-166), and the Mitsubishi Foundation (202210047).
About Professor Masaaki Komatsu, Ph.D. from Juntendo University
Masaaki Komatsu is a Professor in the Department of Organ and Cell Physiology, Juntendo University School of Medicine, Tokyo, Japan. His areas of research interest range from autophagy, the UFMI-system, protein homeostasis, to organelle homeostasis. He has received several awards including the Thomson Reuters Highly Cited Researchers, Japan Academy Medal, JSPS Prize, Young Scientist Award, Young Investigator Award as well as the prestigious Clarivate Analytics Highly Cited Researchers. He has published 168 original articles, 2 book chapters and reviewed 28 articles, and has been cited 43, 417 times according to the Web of Science.
Biomolecular condensates like p62 body have a role in cleansing cellular waste through “autophagy.” Much remains unknown about the constituents of these membrane-less organelles, however. Scientists from Japan have developed a novel method that combines fluorescence-activated particle sorting and selective autophagy-deficient mouse tissues to identify several p62-body components, including a major subunit of the supramolecular protein complex, vault, which is likely linked with non-alcoholic steatohepatitis–derived hepatocellular carcinoma. Their findings are published in Developmental Cell.
Researchers have combined fluorescence-activated particle sorting and selective autophagy-deficient mouse tissues, and identified a novel pathway for selective autophagy
Researchers have combined fluorescence-activated particle sorting and selective autophagy-deficient mouse tissues, and identified a novel pathway for selective autophagy
 The method led to the identification of vault, a component of p62 bodies, with a possible role innon-alcoholic steatohepatitis-derived hepatocellular carcinoma.
The method led to the identification of vault, a component of p62 bodies, with a possible role innon-alcoholic steatohepatitis-derived hepatocellular carcinoma.
Image credits:Masaaki Komatsu from Juntendo University School of Medicine
License type:Original content
Researchers have long been studying the mechanisms related to autophagy and its role in potentially preventing and fighting diseases. p62 protein is an important part of this process. It forms a membrane-less cytosolic organelle called p62 body and helps in the selective removal of toxic cellular waste through autophagy. Regulated removal of unwanted proteins via selective autophagy of p62 bodies maintains cellular homeostasis. However, their compromised clearance leads to the accumulation of p62 and proteins inside p62 bodies, resulting in several diseases, such as hepatocellular carcinoma. While studies have helped us understand the mechanisms associated with the degradation of p62 bodies, our knowledge on their constituents remains incomplete.
To this end, a group of scientists led by Associate Professor Hideaki Morishita and Professor Masaaki Komatsu from Juntendo University Graduate School of Medicine, Japan, have developed a novel method that uses fluorescence-activated particle sorting (FAPS) to purify p62 bodies. They followed it with mass spectrometry of the purified organelles and the tissues retrieved from selective autophagy-defective mice, to identify the substrates within p62 bodies. Their analyses not only identified several previously known autophagy substrates and receptors present within p62 bodies, but also unraveled a major subunit of the supramolecular protein complex, vault, as a cargo within the membrane-less organelle. Their findings have been published in the journal Developmental Cell on May 15, 2023.
The team also included Reo Kurusu and Yuki Fujimoto, undergraduate students from the Program for Training Physicians Conducting Basic Research, Juntendo University. The scientists elaborate that vault is recruited to p62 bodies and degraded by selective autophagy, a process called vault-phagy. This is achieved through an intermediary protein called NBR1, which is critical for the employment of vault and its subsequent degradation along with p62 bodies.
Why is this finding important? According to the scientists, vault-phagy, i.e., the degradation of vault, regulates homeostatic vault levels within cells and tissues, and its impairment may be associated with non-alcoholic steatohepatitis-derived hepatocellular carcinoma, among others.
Discussing their study, Prof. Masaaki remarks, “Our proteomic approach identified the contents of p62 bodies in detail and enabled us to determine the substrates of p62-mediated selective autophagy. This method can be applied to other cell types and tissues under several conditions, like stress and diseases.”
Although there is evidence in the literature on methods to identify selective autophagy substrates, the present method using FAPS and selective autophagy–defective mice has several advantages. FAPS is more suitable for the purification of non-membranous cytosolic organelles like p62 bodies. Furthermore, it can identify pathway-specific substrates and can be instrumental in understanding the physiological role of selective autophagy and phase separation in proteostasis.
As Associate Professor Hideaki Morishita observes, “Our methods have a broad scope of potential applications that should lead to the discovery of currently uncharacterized phase separation–mediated cargoes in order to allow for a deeper understanding of selective autophagy and related diseases.”
While the scientists believe there is a need for further research to understand the implications of vault-phagy in diseases, their novel technique and the insights generated in this study are sure to enrich our knowledge of the mechanisms of our cell’s housekeeping!
Funding
This study is mainly supported by Grant-in-Aid for Scientific Research on Innovative Areas (19H05706), a Grant-in-Aid for Scientific Research (A) (21H004771), Advanced Research and Development Programs for Medical Innovation (AMED-CREST, 22gm1410004s0103), the Takeda Science Foundation, a Grant-in-Aid for Scientific Research (B) (20H03213, 23H02441), Advanced Research and Development Programs for Medical Innovation (AMED-PRIME, 21gm6410019h0001), the Inamori Scientific Foundation (217959), a Kishimoto Fund Research Grant (2022-166), and the Mitsubishi Foundation (202210047).
About Professor Masaaki Komatsu, Ph.D. from Juntendo University
Masaaki Komatsu is a Professor in the Department of Organ and Cell Physiology, Juntendo University School of Medicine, Tokyo, Japan. His areas of research interest range from autophagy, the UFMI-system, protein homeostasis, to organelle homeostasis. He has received several awards including the Thomson Reuters Highly Cited Researchers, Japan Academy Medal, JSPS Prize, Young Scientist Award, Young Investigator Award as well as the prestigious Clarivate Analytics Highly Cited Researchers. He has published 168 original articles, 2 book chapters and reviewed 28 articles, and has been cited 43, 417 times according to the Web of Science.
Reference
| Authors | Reo Kurusu1,9, Yuki Fujimoto1,9, Hideaki Morishita1,9,*, Daisuke Noshiro2, Shuhei Takada1, Koji Yamano3, Hideaki Tanaka4, Ritsuko Arai5, Shun Kageyama1, Tomoko Funakoshi1, Satoko Komatsu-Hirota1, Hikari Taka6, Saiko Kazuno6, Yoshiki Miura6, Masato Koike7, Toshifumi Wakai8, Satoshi Waguri5, Nobuo N Noda2, Masaaki Komatsu1,10,* |
| Title of original paper | Integrated proteomics identifies p62-dependent selective autophagy of the supramolecular vault complex |
| Journal | Developmental Cell |
| DOI | 10.1016/j.devcel.2023.04.015 |
| Affiliations | 1 Department of Physiology, Juntendo University Graduate School of Medicine, Bunkyo-ku, Tokyo 113-8421, Japan
2 Institute for Genetic Medicine, Hokkaido University, Sapporo, 060-0815, Japan 3 Department of Biomolecular Pathogenesis, Medical Research Institute, Tokyo Medical and Dental University, Bunkyo-ku, Tokyo 113-8510, Japan 4 Laboratory for Protein Crystallography, Institute for Protein Research, Osaka University, Suita, Osaka 565-0871, Japan 5 Department of Anatomy and Histology, Fukushima Medical University School of Medicine, Hikarigaoka, Fukushima 960-1295, Japan 6 Laboratory of Proteomics and Biomolecular Science, Biomedical Research Core Facilities, Juntendo University Graduate School of Medicine, Bunkyo-ku, Tokyo 113-8421, Japan 7 Department of Cell Biology and Neuroscience, Juntendo University Graduate School of Medicine, Bunkyo-ku, Tokyo 113-8421, Japan 8 Division of Digestive and General Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata City, Niigata 951-8510 Japan 9 These authors contributed equally 10 Lead contact *Corresponding author |